
Genome-wide association study of knee osteoarthritis: present and future
Introduction
Knee osteoarthritis (KOA) is a common complex disease caused by a combination of genetic and environmental factors. There is strong evidence of genetic influence of OA that comes from many studies, including adoption studies, twin studies and Mendelian disorders studies related to OA. Estimated heritability of OAs is different between OA sites. The heritability of KOA, hip OA and lumber spine OA are 39%, 60% and 74%, respectively (1). Furthermore, both phenotypic and genetic correlations exist in OAs, and it is thought that there is a genetic background common to each OA to some extent. An association study has been used as method to prove the details of genetic factors, and a lot of OA susceptibility genes have been identified until now.
The association study is used to identify disease susceptibility genes of a lot of complex diseases. There are two methods of the association study. One is a candidate gene study that analyzes known genes as candidates. Another is a genome-wide association study (GWAS) that analyzes the whole genome using single nucleotide polymorphisms (SNPs). The GWAS was performed for the first time in the world in 2002 in myocardial infarction (2). With respect to KOA, Nakajima et al. (3) firstly reported in 2010 and there have been a number of GWAS reports to date (Table 1) (5-9).

Full table
In this chapter, we review the current status and problems of genetic studies of KOA and present our approach for early KOA (eKOA) defined.
The current status and problems of genetic studies of KOA
Currently, genetic association studies have identified ~30 independent OA susceptibility loci (10,11). With the exception of GDF5, all loci are identified by GWASs; 10 of them were associated with KOA.
In 2010, Nakajima et al. firstly reported KOA GWAS, which include 899 cases and 3,396 population controls from a Japanese cohort (Table 2) (3). Their disease definition was based on the Kellgren-Lawrence (KL) grade for the knee radiograph [antero-posterior (A-P) view]. After performing replication using 980 cases and 1,418 controls, they identified two KOA susceptibility genes.
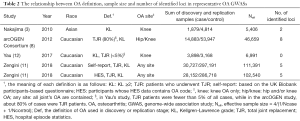
Full table
In 2012, a UK study named arcOGEN performed a GWAS that included 7,410 OA cases (4,144 KOA cases) and 11,009 population controls (8). After performing replication and meta-analysis in up to 14,883 cases and 53,947 controls, they identified eight OA susceptibility loci of genome-wide significance that included four loci of KOA. The characteristic of the UK study was that the definition of OA was different among sub-groups of the study and many of cases using in the discovery stage was the subjects who were regarded as having OA because they just underwent total joint replacement (TJR) surgery irrespective of radiographic data.
In 2017, Yau et al. performed GWAS of KOA in 3,898 cases and 3,168 controls from North American cohorts (12). They used both KL grade and TJR for the definition of OA, but unlike the arcOGEN study, TJR patients were fewer than 5% of all OA cases. Although they unified the interpretation of KL grade 2 among cohorts to reduce the variance of case groups, they could not identify any genome-wide significant loci.
In 2018, Zengini et al. performed an OA GWAS using UK Biobank data (11). In the discovery stage of this study, they used two different OA definitions: self-reported and the Hospital Episode Statistics (HES). After performing replication and meta-analysis in up to 30,727 cases and 297,191 controls, they identified 10 genome-wide significant loci. The characteristic of this study is that they mixed OAs of various sites (hand, spine, hip, knee, and so on) to increase the sample size as much as possible.
KOA susceptibility genes robustly identified by GWAS with sufficient genetic evidence are only nine genes (Table 1), which are far from explaining the heritability of KOA. In addition, there are few genes which satisfied genome-wide significant level of the association in plural GWASs even between the studies of the same ethnic groups. The low replicability could be mainly because low statistical power of these OA GWAS. In GWAS, it is necessary to set a strict standard for correction of the multiple testing [P value ≤5×10−8 =0.05/1,000,000 (the usual upper limit number of the genotyped SNPs in a GWAS)]. Thus, to achieve sufficient statistical power of GWAS, enormous sample is necessary. In other words, OA GWAS’s sample size to date are simply too small to detect many genes. In other complex diseases such as osteoporosis and rheumatoid arthritis, GWASs which included tens of thousands of cases identified more than one hundred susceptibility genes (13,14). Such a powerful GWAS would also identify many KOA susceptibility genes, although to enlarge a sample size is not easy in practice.
Another problem of KOA GWAS is its vague phenotype definition. In many epidemiological and genetic studies, KOA is evaluated by KL grade for the plain knee radiography. Although KL grade 2 or more are usually defined as KOA, KL grade 2 itself is not clearly defined. Kerkhof et al. (15) reported that five different KOA (KL grade 2) definitions were used in 28 studies involved in the TREAT-OA consortium. They revealed that the cause of the difference of the KOA prevalence between studies was the vagueness of the definition and the difference of the prevalence among the studies is decreased by the unification of the KOA definition. Thus, when we use KL grade for defining cases, there are much heterogeneity between studies. This heterogeneity of cases will reduce statistical power of the GWAS analysis.
A distinct phenotype improves the power of OA GWAS
The hip OA GWAS conducted by Castaño-Betancourt et al. (16) in 2016 is a good example to highlight the importance of the phenotype definition. They used a minimal joint space width (mJSW) based on the plain hip radiography (A-P view) as a proxy for cartilage thickness. Then, GWAS of mJSW was performed in a discovery set that included a modest sample size (13,013 individuals). In this discovery stage, four genetic loci met the genome-wide significance threshold. By contrast, in UK Biobank OA GWAS (11) using HES data, no locus met the genome-wide significance threshold in the discovery stage although its effective sample size (n=32,280) was quite large. HES is a database containing details of all admissions and outpatient appointments at National Health Service hospitals in England. Thus, the data of HES are a collection of disease names diagnosed by many doctors (specialists) in daily practice. Specialists diagnose OA by KL grade (a combination of bone and/or cartilage features) as well as clinical complaints in daily practice and this vague criterion causes the heterogeneity of the HES date. On the other hand, mJSW focus on the only cartilage thickness, which is the main affected tissue of OA and is more objective. Furthermore, a quantitative trait like mJSW has more information than a binary trait. We suspect that this phenotype difference may cause the different statistical power between these two GWAS.
Fusion of eKOA study and OA genetic studies
In recent years, the concept of eKOA for the early detection and treatment of KOA has been proposed, and ESSKA (European Society of Sports Traumatology Knee surgery and Arthroscopy) published its definition in 2012 (17). Before being able to be caught as a joint space narrowing in the plain knee radiography, cartilage damage exists, which is detected by MRI or arthroscopy and when it accompanies knee pain, it is defined as eKOA. Thus, this definition evaluates the change of cartilage, the main affected tissue of OA more strictly. The clinical significance of this definition is still unknown, and at the moment there is also no genetic study related to eKOA. However, we believe that the idea of detecting minute changes in cartilage, which is the earliest change of OA, and making the definition strict is a concept applicable to phenotypic determination in genetic study.
We had started working on eKOA research based on detecting early changes of cartilage before the definition of ESSKA was advocated. Invasive and costly inspection methods such as MRI and arthroscopy are unsuitable for genetic studies which require enormous sample sizes and we adopted US evaluation which is simpler and cheaper. Since 2011 we are conducting cohort studies based on this classification. (see Uchio and Kumahashi’s chapters). We conducted an epidemiological research based on our US classification and clarified the distribution and epidemiological features of KOA defined by US of the general population. Furthermore, we showed that individuals whose KL grade 0 or 1 could be subdivide by the US classification. We hope that the distinct phenotype definition based on our US evaluation will improve the KOA GWAS. We have already started a GWAS using our cohort data in RIKEN.
To reveal the etiology and pathogenesis of KOA, it is necessary to approach them using the knowledge of orthopaedics and genome medicine. The more distinct definition of KOA established with accumulation of the evidence of eKOA will improve the power of GWASs. The more powerful GWAS will identify more KOA susceptibility genes, which will lead to elucidate the pathogenesis of KOA and establish the effective cure and prophylaxis.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Joint for the series “Early Osteoarthritis: Definition, Pathogenesis, Diagnosis, Management and Prevention”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aoj.2018.07.04). The series “Early Osteoarthritis: Definition, Pathogenesis, Diagnosis, Management and Prevention” was commissioned by the editorial office without any funding or sponsorship. YU served as the unpaid Guest Editor of the series. SI serves as an Editor-in-Chief of Annals of Joint from Mar 2016 to Feb 2021.The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 2004;12 Suppl A:S39-44.
- Ozaki K, Ohnishi Y, Iida A, et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 2002;32:650-4. [Crossref] [PubMed]
- Nakajima M, Takahashi A, Kou I, et al. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One 2010;5:e9723 [Crossref] [PubMed]
- Miyamoto Y, Mabuchi A, Shi D, et al. A functional polymorphism in the 5' UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 2007;39:529-33. [Crossref] [PubMed]
- Miyamoto Y, Shi D, Nakajima M, et al. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat Genet 2008;40:994-8. [Crossref] [PubMed]
- Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis 2011;70:349-55. [Crossref] [PubMed]
- Day-Williams AG, Southam L, Panoutsopoulou K, et al. A variant in MCF2L is associated with osteoarthritis. Am J Hum Genet 2011;89:446-50. [Crossref] [PubMed]
- arcOGEN Consortium; arcOGEN Collaborators, Zeggini E, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 2012;380:815-23.
- Valdes AM, Evangelou E, Kerkhof HJ, et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis 2011;70:873-5. [Crossref] [PubMed]
- Cibrián Uhalte E, Wilkinson JM, Southam L, et al. understanding the genomic aetiology of osteoarthritis. Hum Mol Genet 2017;26:R193-201. [Crossref] [PubMed]
- Zengini E, Hatzikotoulas K, Tachmazidou I, et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet 2018;50:549-58. [Crossref] [PubMed]
- Yau MS, Yerges-Armstrong LM, Liu Y, et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol 2017;69:343-51. [Crossref] [PubMed]
- Karasik D, Rivadeneira F, Johnson ML. The genetics of bone mass and susceptibility to bone diseases. Nat Rev Rheumatol 2016;12:323-34. [Crossref] [PubMed]
- Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506:376-81. [Crossref] [PubMed]
- Kerkhof HJ, Meulenbelt I, Akune T, et al. Recommendations for standardization and phenotype definitions in genetic studies of osteoarthritis: the TREAT-OA consortium. Osteoarthritis Cartilage 2011;19:254-64. [Crossref] [PubMed]
- Castaño-Betancourt MC, Evans DS, Ramos YF, et al. Novel Genetic Variants for Cartilage Thickness and Hip Osteoarthritis. PLoS Genet 2016;12:e1006260 [Crossref] [PubMed]
- Luyten FP, Denti M, Filardo G, et al. Definition and classification of early osteoarthritis of the knee. Knee Surg Sports Traumatol Arthrosc 2012;20:401-6. [Crossref] [PubMed]
Cite this article as: Takuwa H, Uchio Y, Ikegawa S. Genome-wide association study of knee osteoarthritis: present and future. Ann Joint 2018;3:64.


