
Advances in understanding the genetics of syndromes involving congenital upper limb anomalies
Introduction
Congenital upper limb anomalies (CULA) include a wide spectrum of structural abnormalities caused by perturbation in the morphogenesis of the upper limb, which affect approximately 1 per 500 live births (1,2). About 20% to 30% of babies with CULA has at least one associated non-limb anomaly (2), and approximately 10% to 20% of all congenital anomalies has upper limb involvement (3). This condition exhibits an extreme heterogeneity of presentations with different grades of severity. Although some of them result in mild functional consequences, they can also bring psychological problems for children (4).
So far, there are two commonly used classification schemes in clinical settings, i.e., the Swanson Classification and the Oberg-Manske-Tonkin (OMT) Classification. The Swanson Classification was proposed in 1964 and utilized the anatomical understanding and the surgical perspective on management (5). Although the Swanson Classification is satisfactory in several criteria, it does not include the knowledge acquired in the last 50 years (2). The OMT Classification combines more recent knowledge about limb development and the pathogenesis of limb anomalies using dysmorphology terminology (6) and provides a more practical and easily utilized scheme.
The pathogenesis of CULA remains poorly understood. A limited number of studies provide evidence in linking exposure to environmental chemicals and CULA, including persistent organic and air pollution (7). Interestingly, chorionic villus sampling has been identified as a risk factor of syndromic and non-syndromic CULA (8). In contrast, the observation of familial CULA cases highlighted the contribution of genetic factors in CULA. Novel approaches such as single nucleotide polymorphism (SNP) array, comparative genomic hybridization-array (array-CGH) and next generation sequencing (NGS) enable the identification of numerous causative genes of CULA. Unravelling the genetic background of syndromic CULA helps to broaden the spectrum of causative genes, identify the pathways required for normal limb organogenesis and explain the mechanism underlying the cooccurrence of CULA and non-limb anomalies. In this review, we first introduce the developmental process of the upper limbs and the involved signaling pathways. Then, we provide an overview of the genetic basis of syndromic CULA according to the related pathways of the causal genes.
Development of the upper limbs
The development of vertebrate limbs during embryonic life is a multi-stage process, which is orchestrated by complex interactions between signaling centers. In humans, the development of the limb bud initiates at around the fourth week of gestation with the appearance of a small bud from the lateral body wall. The limb bud consists of an inner mesenchymal core and an outer ectodermal cap (9). Following the limb bud emergence is the formation of three axes: the proximal-distal (PD), the anteroposterior (AP) and the dorso-ventral (DV) axes. Mesenchymal fibroblast growth factor 10 (FGF10), bone morphogenetic proteins (BMP) signaling induce the formation of the apical ectodermal ridge (AER), which is located at the DV interface. AER mainly mediates PD patterning through the regulation of FGF and WNT signals (10). Besides FGF from AER, DV axis is also defined by retinoic acid (RA) signaling from the lateral walls (9). Zone of polarizing activity (ZPA) locates in the posterior limb bud mesenchyme and is controlled by Sonic Hedgehog (SHH) signaling. SHH secreted by ZPA diffuse along the limb bud to form a gradient, which control the formation of AP axis. Multiple signals including TBX, HAND2, RA, HOXD and GLI3R are involved in the regulation of SHH expression and further influence AP patterning. SHH from ZPA and FGF from AER form an epithelial-mesenchymal feedback loop that governs the formation of PD and AP axis. WNT family member 7A (WNT7A), produced by the presumptive dorsal limb ectoderm, is considered to be the organizer of DV axis. The specific expression of LIM-homeodomain transcription factor 1 beta (LMX1B) in the dorsal mesenchyme of the limb bud is induced by WNT7A (6). LMX1B is a necessary and sufficient factor in guidance of the dorsal limb patterning (11). The signal transduction of SHH, WNT and BMP is critically coordinated by the primary cilium, which is a microtubule-based organelle that emerge from the cell surface of most vertebrate cell types (10).
Genetic advances in syndromic CULA
Aat)Non-limb anomalies associated with syndromic CULA include a broad range of presentations, e.g., genitourinary, cardiovascular, nervous, facial, etc. (11). Syndromic CULA contribute to 20% to 30% of all CULA (1,2), and about 10% to 20% of all congenital anomalies have upper limb involvement (3). Associated non-limb anomalies can be life threatening and thus require more attention from clinicians and parents.
Both genetic mutations and genomic rearrangements can lead to syndromic CULA (summarized in Table 1). Many genes involved in the pathogenesis of syndromic CULA can be attributed to several biological pathways. Here, we introduce roles of Cohesin proteins, WNT signaling, Hedgehog signaling, FGFs, FGFRs and PI3K-AKT1-mTOR signaling in the pathogenesis of syndromic CULA.
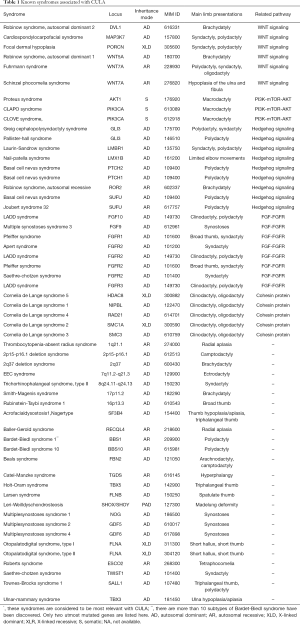
Full table
Cohesin proteins and Cornelia de Lange syndrome
Cohesin proteins are important Structural Maintenance of Chromosomes (SMC) protein containing complexes. By mediating the chromatin organization and long-distance chromatin interaction, the cohesion proteins can affect gene expression (12). Several disorders originate from mutations in cohesin subunits and its key regulators are termed as cohesinopathies. Cornelia de Lange syndrome (CdLS; MIM: 122470), a prominent member of cohesinopathies, is a dominantly inherited disorder characterized by dysmorphic features, CULA, global developmental retardation and gastrointestinal system involvement. NIPBL load the cohesin complex onto DNA and is required for topological movements of DNA (13). Mutations in NIPBL are responsible for a significant proportion of CdLS (12). Currently there are six cohesins-related genes have been implicated in CdLS, i.e., HDAC8 (MIM: 300882), RAD21 (MIM: 614701), NIPBL (MIM: 122470), SMC1A (MIM: 300590), BRD4 and SMC3 (MIM: 610759) (14). Among them, HDAC8, BRD4 and NIPBL are key regulators of cohesin proteins while SMC1A. SMC3 and RAD21 are structural components of the cohesin ring (13,15).
NIPBL induce histone deacetylation through recruiting histone deacetylase (HDAC) and regulate gene expression. In a zebrafish model with Nipbl knockdown, the size reductions and patterning defects of pectoral fin (forelimb) were observed along with impaired expression of several key limb development genes including fgfs, hand2 and multiple hox genes (16), which suggests that the limb malformations in CdLS can be attributed to the impact of Cohesin proteins upon the expression of critically limb development genes.
WNT signaling
WNT signals comprise a diverse group of secreted proteins that mediates crucial developmental processes including embryonic axis patterning, cell fate specification, proliferation and migration (17). Genes coding for a variety of signaling molecules involved in Wnt signaling pathway have been identified in Mendelian inherited syndromic CULA. Recessively inherited WNT7A mutations were described in individuals with Schinzel phocomelia syndrome (MIM: 276820) and Fuhrmann syndrome (MIM: 228930) (18). Both syndromes have limb deformities, but Schinzel phocomelia syndrome comprise a broader set of phenotypes including severe anomalies of upper and lower limbs with hypoplastic pelvis and genitalia. There are many other genes in Wnt signaling pathway which have been associated with syndromic CULA. Cardiospondylocarpofacial syndrome (CSCF; MIM: e157800) is characterized by cardiac septal defects, vertebral synostosis, brachydactyly with carpal-tarsal fusion and dysmorphic facial features. CSCF is caused by heterozygous mutation in MAP3K7 gene. MAP3K7 act as an upstream kinase regulating several pathways including WNT, NF-κB and p38 MAPK (19). Focal dermal hypoplasia (MIM: 305600), caused by mutations in PORCN, is a X-linked dominant syndrome with in utero lethality in males. Common features of this syndrome include distinct skin manifestations, digits malformations (mainly comprise syndactyly and polydactyly), oral and ocular anomalies. PORCN is essential for the acetylation and secretion of WNT ligands in mice and humans (20). Given the crucial role of Wnt signaling in human development, it is not surprised to find out these Wnt signaling associated syndromic CULA display complex phenotypes commonly affecting multiple organs and systems.
Robinow syndrome is a genetically heterogeneous severe skeletal dysplasia characterized by mesomelic limb shortening, dysmorphic features including midface hypoplasia, hypoplastic genitalia and vertebral anomalies (21,22). Mutations of several genes involved in the WNT signaling pathway have been implicated in different subtypes of Robinow syndrome. WNT5A is involved in both the canonical and noncanonical Wnt signaling pathway (23). Dominantly inherited WNT5A mutations have been associated with autosomal dominant Robinow syndrome-1 (DRS; MIM: 180700) and homozygous Wnt5a null mice presented with anatomical defects resembling individuals with autosomal DRS (24). ROR2, a tyrosine kinase-like orphan receptor, interacts with WNT5A both functionally and physically (23). The binding of WNT5A with ROR2 lead to phosphorylation of ROR2, resulting in Rho GTPase dependent activation of the WNT-JNK and WNT calcium pathway, i.e., the noncanonical WNT pathway. Recessive and dominant mutations of ROR2 have also been implicated in autosomal recessive Robinow syndrome (RRS; MIM: 268310) and brachydactyly type B1 (BDB1; MIM: 113000), respectively. Interestingly, not all patients carrying heterozygous null mutations in ROR2 exhibited brachydactyly. By measuring steady-state protein model levels as well as surface location in ROR2 variant-expressing cell lines, Schwarzer et al. presented a hypothesis that the phenotypic outcome can be explained by the equilibrium between intracellular retention and cell surface expression rather than a simple loss-or gain-of-function model (25). Dishevelled (DVL): 13 is intracellular protein Tinvolved in the canonical and non-canonical WNT signaling (26). The binding of DVL to the cell membrane facilitates the binding of Axin and GSK3β, which subsequently phosphorylate LRP5/6 and thereby preventing degradation of β-catenin. The accumulation of β-catenin in the nucleus activates WNT responsive genes (26). In the non-canonical WNT pathway, DVL plays key role in regulating polarity and cytoskeletal determination by acting as a branchpoint for activation of Rho, Rac and Cdc42 (26). Heterozygous DVL1 and DVL3 mutations have been shown to lead to autosomal dominant Robinow syndrome-2 (MIM: 616331) and autosomal dominant Robinow syndrome-3 (MIM: 616894) (27). Together, these evidences suggest that WNT signaling pathway play an important role in the pathogenesis of a wide spectrum of hypoplastic disorders including CULA.
Hedgehog signaling
The Hedgehog (HH) signaling pathway has many roles in controlling of the cell proliferation, embryonic patterning and limb development (9). Gli-kruppel (GLI) genes encode transcription factors involved in SHH signaling pathway. GLI1, GLI2 and GLI3, members of GLI family, have been associated with multiple human diseases including both syndromic and non-syndromic CULA. GLI3 is a transcription factor and act as a modulator for SHH pathway with either facilitative and suppressive function (28). SHH pathway inactivation constitutively transforms GLI3 to GLI3R isoform, which is translocated into the nucleus and negatively regulates SHH target genes (29). Activated SHH inhibits GLI3R isoform formation and induces the formation of GLI2A and GLI3A (GLI activators). Subsequently, GLI activators promotes the expression of SHH targeted genes. Besides ROR2, GLI3 is another gene that have been implicated in both syndromic and non-syndromic CULA. Heterozygous mutations in GLI3 gene can lead to Greig cephalopolysyndactyly syndrome (GCPS; MIM: 175700), Pallister-Hall syndrome (PHS; MIM: 146510), postaxial polydactyly type A1 and B (PAP; MIM:174200) and preaxial polydactyly type IV (PPD; MIM: 174700) (28). GCPAS and PHS are allelic syndromes with distinct clinical entities (30). GCPS is mainly characterized by polysyndactyly, macrocephaly with frontal bossing and hypertelorism (31). Preaxial of the feet and postaxial in the hands are most commonly identified forms of polydactyly (31). Typical phenotypes of PHS include central polydactyly, pituitary malfunction, hypothalamic hamartoma and visceral malformations (30). The genotype-phenotype correlations of GLI3 associated disorders are well characterized (28). The N-terminal part of the GLI3 protein contains the zinc finger domain. Mutations in the zinc finger domain mostly result in GCPS through a haplo-insufficiency mechanism. The middle part of the GLI3 protein encompasses the protein cleavage site. Patients with PHS usually carry mutations in the middle part. The C-terminal of contains the transactivating domains 1 and 2. Mutations in the C-terminal part lead to distinct phenotypes including GCPS, PAP and PPD (28). Nevertheless, the mechanism underlying the distinct phenotype spectrum needs further studies.
SUFU, acronym for Suppressor of Fused, plays a critical role in primary cilia. SUFU acts as the main negative regulator of the SHH pathway through forming a complex with GLI3 and GLI2 in primary cilia (32). Transportation of SUFU to primary cilia is potentially governed by GLI activators but not suppressors (33). Recessive and dominant mutations of SUFU were known to cause Joubert syndrome 32 (MIM: 617757) and basal cell nevus syndrome (BCNS; MIM: 109400), respectively (32). Joubert syndrome 32 is characterized by congenital ataxia, post-axial polydactyly, cerebellar vermis hypoplasia and cranio-facial dysmorphisms. While postaxial syndactyly is relatively rare in BCNS (32). Heterozygous mutations in PTCH1 and PTHC2 can also lead to basal cell nevus syndrome. Basal cell nevus syndrome is a familial cancer predisposing syndrome which comprises a broad spectrum of systemic manifestations, the most characteristic symptoms of which are basal cell carcinomas, jaw keratocytes and cerebral calcifications (34). PTCH1 and PTCH2, encoding members of the patched family, suppress SHH signaling through inhibition of SMO. Loss-of-function of PTCH1 and PTCH2 result in aberrant increase in SHH signaling (34).
The ZPA regulatory sequence (ZRS), a long-range limb-specific enhancer of the SHH, is located in intron 5 of LMBR1 (35). Recently, a missense mutation in p-ZRS (pre-ZRS), a noncoding sequence 500 bp upstream of the ZRS, have been associated with triphalangeal thumb-polysyndactyly syndrome (MIM: 174500) in a multigenerational family (36). Transgenic mice carrying this mutation showed ectopic SHH expression (36). Although regulatory effects of ZRS upon Shh have been well characterized (37), further study is still needed to determine the role of pZRS in Shh limb expression.
Fibroblast growth factors (FGFs) and FGFRs
FGFs are a family of proteins that regulates many developmental processes in the early stage of embryonic development. FGFs carry out their functions through binding to the fibroblast growth factor receptors (FGFRs). Currently there are four FGFRs (FGFR1 to FGFR4) known to interact with FGFs in an HSGAG-dependent manner (38). Either increasing and decreasing of FGF signaling can cause human diseases. For example, gain-of-function mutations of FGFR2 lead to Apert syndrome (MIM: 101200) (39), a developmental disorder characterized by craniosynostosis, midface hypoplasia and severe syndactyly with fusion tendency. Studies have revealed that about two-thirds of Apert syndrome is caused by the S252W mutation in FGFR2, and the rest one third is caused by the P253R mutation in FGFR2 (40). Syndactyly in hands and feet was likely to be more severe in patients carry the P253R mutation, while cleft palate was more frequently observed in patients carrying the P252W mutation (40). Prolyl isomerase peptidyl-prolyl cis–trans isomerase interacting 1 (PIN1) is a crucial regulator of FGFR signaling and Pin1+/− mice displayed delayed closure of cranial sutures (41). By crossing Pin1+/− mice with Fgfr2S252W/+ mice, a mouse model of Apert syndrome, the downregulation of Pin1 function attenuated premature cranial and frontal-nasal suture fusion. Although other phenotypes like syndactyly was not rescued by the reduced dosage of Pin1, this approach do provide novel therapeutic insights in alleviating other phenotypes in Apert syndrome. Gain-of-function mutations of FGF10 is responsible for Lacrimoauriculodentodigital syndrome (LADD syndrome; MIM: 149730) (42), a congenital disorder mainly involving lacrimal glands and duct, ears, teeth and digits. Activating mutations in FGFR2 and FGFR3 can also lead to LADD syndrome (43). FGFR2 interacts with FGF10 and conditional knockout of Fgfr2 in mice leads to limb and digit malformations (43). Mutations in FGFR3 gene are now associated with at least10 human disorders, involving skeletal dysplasia, skin malformation and cancer (44). Attenuation of the chondrocytes are responsible for the FGFR3-related skeletal dysplasia (44). Recently, Bosakova et al., found that the sustained and transient activation of FGF signaling result in shortening and elongation of primary cilia, respectively (45). The SHH signaling is subsequently influenced by the impaired cilia function (45), providing insights into the pathogenic mechanism underlying the limb phenotypes of FGFR-related disorders.
PI3K-AKT1-mTOR
PI3K-AKT-mTOR signaling mediates cell proliferation, survival and metabolism, activating of this pathway is commonly involved in tumorigenesis. PIK3CA and AKT1 are frequently mutated gene in human tumors (46,47). Somatic mosaicism of PIK3CA and AKT1 can both lead to overgrowth syndromes. Genetic mosaicism refers to a process occurring postzygotically that result in the presence of genetically distinct cells within one individual. Advent of the deep NGS facilitated the discovery of developmental disorders caused by mosaic mutations. Proteus syndrome (MIM: 176920), caused by mutations in AKT1, is characterized by cerebriform connective tissue nevi and patchy overgrowth most commonly involving limbs. Most patients with Proteus syndrome carried a hotspot gain of function mutation c.G49A in AKT1 (47). Somatic mosaicism of PIK3CA is responsible for a wide spectrum of disorders named as PIK3CA related overgrowth syndrome, including congenital lipomatous overgrowth, vascular malformations, epidermal nevi and skeletal/scoliosis/spinal abnormalities (CLOVE syndrome; MIM: 612918), capillary malformation of the lower lip, lymphatic malformation of the face and neck, asymmetry and partial/generalized overgrowth (CLAPO syndrome; MIM: 613089), megalencephaly-capillary malformation (MCAP, MIM: 602501). Focal forms of PIK3CA related overgrowth include macrodactyly, epidermal nevi, infiltrating lipomatosis lymphatic malformations and venous malformations (48). The most frequently observed limb component of these disorders is macrodactyly and the degree of overgrowth is variable.
CNVs
Several well-characterized CNVs underlying syndromes with limb involvement have been identified. Typically, thrombocytopenia-absent radius syndrome (TAR syndrome; MIM: 274000), a syndrome with multiple anomalies affecting the blood circulation of the upper limb, is caused by compound heterozygosity for a 1q21.1 deletion involving the RBM8A gene and a noncoding SNP in RBM8A (49). The impact of a rare null variant and a noncoding common SNP was also observed in TBX6-associated congenital scoliosis (TACS) (50,51). A common TBX6 hypomorphic allele in trans with a rare 16p11.2 deletion or TBX6 loss-of-function variant lead to TACS (52). Mouse models of TBX6 compound heterozygosity display vertebral malformations and provide further evidence supporting the gene dosage model (51,52). Blood system anomalies encompass reduction in the number of platelets in blood and reduced number of megakaryocytes and platelet precursor cells in bone marrow. The severity of the upper limb involvement ranges from the absence of the radius to the absence of the most part of upper limbs, but thumbs are always well-preserved. Malformations in spine, lower limbs are less common.
Another well-characterized CULA associated syndrome is 2p15-p16.1 microdeletion syndrome (MIM: 612513) (53). This syndrome has a wide spectrum of phenotypes and about 80% patients present with hand anomalies, mainly consisting of camptodactyly (54). The size of the deletions (ranging from 0.1 to 9.5 Mb) and breakpoints vary dramatically. Recently, XPO1, REL and BCL11A have been identified as candidate genes for 2p15-p16.1 microdeletion syndrome (54).
There are other recurrent CNVs associated with syndromic CULA, e.g., including 2q37 (MIM: 600430), 7q11.2-q21.3 (MIM: 129900), 16p13.3 (MIM: 613458) and 17p11.2 (MIM: 182290). In a large cohort of individuals with non-syndromic congenital limb malformation, high resolution CNV analysis identified that 10% individuals harbored disease relevant CNVs (55). Among CNVs identified in that study, 57% affect the noncoding cis-regulatory genome, suggesting the contribution of non-coding regions to non-syndromic CULA.
Conclusions
The last decade has witnessed dramatic progress in the genetic and genomic sequencing technologies. With the improvements in molecular genetics in past few years, the field of CULA genetics is progressing very rapidly. These improvements have allowed a detailed genotype-phenotype and genotype-prognosis correlation for CULA-associated syndromes. More precise diagnosis and managements can be adopted utilizing this knowledge. Moreover, the association between limb phenotypes and specific genetic/genomic variants has led to the identification of genes indispensable in the limb development and subsequently to a better understanding of the mechanisms regulating limb development. This knowledge gained from syndromes with CULA will help us decipher the genetic basis of CULA also in non-syndromic individuals.
Acknowledgments
Funding: This study was funded in part by the National Key Research and Development Program of China (No. 2016YFC0901501), National Natural Science Foundation of China (81822030, 81772299), Beijing JST Research Funding 2019-YJ03, Beijing Jishuitan Hospital Nova Program XKXX201818 and the CAMS Initiative Fund for Medical Sciences (2016-I2M-3-003, 2016-I2M-2-006, 2017-I2M-2-001).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aoj.2019.06.03). NW serves as an unpaid editorial board member of Annals of Joint from Aug 2018 to Jul 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Giele H, Giele C, Bower C, et al. The incidence and epidemiology of congenital upper limb anomalies: A total population study. J Hand Surg Am 2001;26:628-34. [Crossref] [PubMed]
- Ekblom AG, Laurell T, Arner M. Epidemiology of Congenital Upper Limb Anomalies in Stockholm, Sweden, 1997 to 2007: Application of the Oberg, Manske, and Tonkin Classification. J Hand Surg Am 2014;39:237-48. [Crossref] [PubMed]
- Alrabai HM, Farr A, Bettelheim D, et al. Prenatal diagnosis of congenital upper limb differences: a current concept review. J Matern Fetal Neonatal Med 2017;30:2557-63. [Crossref] [PubMed]
- Andersson GB, Gillberg C, Fernell E, et al. Children with surgically corrected hand deformities and upper limb deficiencies: self-concept and psychological well-being. J Hand Surg Eur Vol 2011;36:795-801. [Crossref] [PubMed]
- Swanson AB. A classification for congenital malformations of the hand. N J Bull Acad Med 1964;10:166.
- Oberg KC, Feenstra JM, Manske PR, et al. Developmental Biology and Classification of Congenital Anomalies of the Hand and Upper Extremity. J Hand Surg Am 2010;35:2066-76. [Crossref] [PubMed]
- Foster WG, Evans JA, Little J, et al. Human exposure to environmental contaminants and congenital anomalies: a critical review. Crit Rev Toxicol 2017;47:59-84. [Crossref] [PubMed]
- Froster UG, Baird PA. Limb-reduction defects and chorionic villus sampling. Lancet 1992;339:66. [Crossref] [PubMed]
- Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet 2009;10:845-58. [Crossref] [PubMed]
- Anvarian Z, Mykytyn K, Mukhopadhyay S, et al. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol 2019;15:199. [Crossref] [PubMed]
- Tonkin MA. Classification of congenital anomalies of the hand and upper limb. J Hand Surg Eur Vol 2017;42:448-56. [Crossref] [PubMed]
- Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Curr Opin Genet Dev 2016;37:59-66. [Crossref] [PubMed]
- Olley G, Ansari M, Bengani H, et al. BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange–like syndrome. Nat Genet 2018;50:329. [Crossref] [PubMed]
- Yuan B, Neira J, Pehlivan D, et al. Clinical exome sequencing reveals locus heterogeneity and phenotypic variability of cohesinopathies. Genet Med 2019;21:663-75. [Crossref] [PubMed]
- Deardorff MA, Wilde JJ, Albrecht M, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet 2012;90:1014-27. [Crossref] [PubMed]
- Muto A, Ikeda S, Lopez-Burks ME, et al. Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development. PLoS Genet 2014;10:e1004671 [Crossref] [PubMed]
- Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017;169:985-99. [Crossref] [PubMed]
- Woods CG, Stricker S, Seemann P, et al. Mutations in WNT7A cause a range of limb malformations, including Fuhrmann syndrome and Al-Awadi/Raas-Rothschild/Schinzel phocomelia syndrome. Am J Hum Genet 2006;79:402-8. [Crossref] [PubMed]
- Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ 2014;21:1667-76. [Crossref] [PubMed]
- Biechele S, Cockburn K, Lanner F, et al. Porcn-dependent Wnt signaling is not required prior to mouse gastrulation. Development 2013;140:2961-71. [Crossref] [PubMed]
- White J, Mazzeu JF, Hoischen A, et al. DVL1 Frameshift Mutations Clustering in the Penultimate Exon Cause Autosomal-Dominant Robinow Syndrome. Am J Hum Genet 2015;96:612-22. [Crossref] [PubMed]
- White JJ, Mazzeu JF, Hoischen A, et al. DVL3 Alleles Resulting in a −1 Frameshift of the Last Exon Mediate Autosomal-Dominant Robinow Syndrome. Am J Hum Genet 2016;98:553-61. [Crossref] [PubMed]
- Person AD, Beiraghi S, Sieben CM, et al. WNT5A mutations in patients with autosomal dominant Robinow syndrome. Dev Dyn 2010;239:327-37. [PubMed]
- Oishi I, Suzuki H, Onishi N, et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells 2003;8:645-54. [Crossref] [PubMed]
- Schwarzer W, Witte F, Rajab A, et al. A gradient of ROR2 protein stability and membrane localization confers brachydactyly type B or Robinow syndrome phenotypes. Hum Mol Genet 2009;18:4013-21. [Crossref] [PubMed]
- Sharma M, Castro-Piedras I, Simmons GE, et al. Dishevelled: A masterful conductor of complex Wnt signals. Cell Signal 2018;47:52-64. [Crossref] [PubMed]
- White JJ, Mazzeu JF, Coban-Akdemir Z, et al. WNT Signaling Perturbations Underlie the Genetic Heterogeneity of Robinow Syndrome. Am J Hum Genet 2018;102:27-43. [Crossref] [PubMed]
- Démurger F, Ichkou A, Mougou-Zerelli S, et al. New insights into genotype–phenotype correlation for GLI3 mutations. Eur J Hum Genet 2015;23:92-102. [Crossref] [PubMed]
- Lopez-Rios J. The many lives of SHH in limb development and evolution. Semin Cell Dev Biol 2016;49:116-24. [Crossref] [PubMed]
- Johnston JJ, Olivos-Glander I, Killoran C, et al. Molecular and Clinical Analyses of Greig Cephalopolysyndactyly and Pallister-Hall Syndromes: Robust Phenotype Prediction from the Type and Position of GLI3 Mutations. Am J Hum Genet 2005;76:609-22. [Crossref] [PubMed]
- Biesecker LG. The Greig cephalopolysyndactyly syndrome. Orphanet J Rare Dis 2008;3:10. [Crossref] [PubMed]
- De Mori R, Romani M, D’Arrigo S, et al. Hypomorphic Recessive Variants in SUFU Impair the Sonic Hedgehog Pathway and Cause Joubert Syndrome with Cranio-facial and Skeletal Defects. Am J Hum Genet 2017;101:552-63. [Crossref] [PubMed]
- Zhang Z, Shen L, Law K, et al. Suppressor of Fused Chaperones Gli Proteins To Generate Transcriptional Responses to Sonic Hedgehog Signaling. Mol Cell Biol 2017;37:e00421-16. [Crossref] [PubMed]
- John AM, Schwartz RA. Basal cell naevus syndrome: an update on genetics and treatment. Br J Dermatol 2016;174:68-76. [Crossref] [PubMed]
- Furniss D, Lettice LA, Taylor IB, et al. A variant in the sonic hedgehog regulatory sequence (ZRS) is associated with triphalangeal thumb and deregulates expression in the developing limb. Hum Mol Genet 2008;17:2417-23. [Crossref] [PubMed]
- Potuijt JW, Baas M, Sukenik-Halevy R, et al. A point mutation in the pre-ZRS disrupts sonic hedgehog expression in the limb bud and results in triphalangeal thumb–polysyndactyly syndrome. Genet Med 2018;20:1405-13. [Crossref] [PubMed]
- Sagai T, Hosoya M, Mizushina Y, et al. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 2005;132:797-803. [Crossref] [PubMed]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 2009;8:235-53. [Crossref] [PubMed]
- Yu K, Herr AB, Waksman G, et al. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci 2000;97:14536-41. [Crossref] [PubMed]
- Slaney SF, Oldridge M, Hurst JA, et al. Differential effects of FGFR2 mutations on syndactyly and cleft palate in Apert syndrome. Am J Hum Genet 1996;58:923-32. [PubMed]
- Shin HR, Bae HS, Kim BS, et al. PIN1 is a new therapeutic target of craniosynostosis. Hum Mol Genet 2018;27:3827-39. [PubMed]
- Milunsky JM, Zhao G, Maher T, et al. LADD syndrome is caused by FGF10 mutations. Clin Genet 2006;69:349-54. [Crossref] [PubMed]
- Rohmann E, Brunner HG, Kayserili H, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet 2006;38:414-7. [Crossref] [PubMed]
- Foldynova-Trantirkova S, Wilcox WR, Krejci P. Sixteen years and counting: the current understanding of fibroblast growth factor receptor 3 (FGFR3) signaling in skeletal dysplasias. Hum Mutat 2012;33:29-41. [Crossref] [PubMed]
- Kunova Bosakova M, Varecha M, Hampl M, et al. Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum Mol Genet 2018;27:1093-105. [Crossref] [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science 2004;304:554. [Crossref] [PubMed]
- Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med 2011;365:611-9. [Crossref] [PubMed]
- Mirzaa G, Timms AE, Conti V, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight 2016;1: [Crossref] [PubMed]
- Albers CA, Paul DS, Schulze H, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet 2012;44:435-9. [Crossref] [PubMed]
- Wu N, Ming X, Xiao J, et al. TBX6 Null Variants and a Common Hypomorphic Allele in Congenital Scoliosis. N Engl J Med 2015;372:341-50. [Crossref] [PubMed]
- Liu J, Wu N, Yang N, et al. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet Med 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Liu J, Zhou Y, Liu S, et al. The coexistence of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) at a locus can result in distorted calculations of the significance in associating SNPs to disease. Hum Genet 2018;137:553-67. [Crossref] [PubMed]
- de Leeuw N, Pfundt R, Koolen DA, et al. A newly recognised microdeletion syndrome involving 2p15p16.1: narrowing down the critical region by adding another patient detected by genome wide tiling path array comparative genomic hybridisation analysis. J Med Genet 2008;45:122-4. [Crossref] [PubMed]
- Bagheri H, Badduke C, Qiao Y, et al. Identifying candidate genes for 2p15p16.1 microdeletion syndrome using clinical, genomic, and functional analysis. JCI Insight 2016;1:e85461 [Crossref] [PubMed]
- Flöttmann R, Kragesteen BK, Geuer S, et al. Noncoding copy-number variations are associated with congenital limb malformation. Genet Med 2018;20:599-607. [Crossref] [PubMed]
Cite this article as: Sun L, Huang Y, Zhao S, Zhong W, Lin M, Guo Y, Yin Y, Wu N, Wu Z, Tian W. Advances in understanding the genetics of syndromes involving congenital upper limb anomalies. Ann Joint 2019;4:30.


