
Autophagy markers and RNA-dependent protein kinase (PKR) activity in osteosarcoma diagnosis and treatment
Introduction
Osteosarcoma is the most common primary bone tumor that mainly affects children and young adults (1,2). The prognosis for osteosarcoma patients still remains poor. Although the overall incidence is low, about 30% of osteosarcoma patients develop metastasis (2). Surgical resection and adjunctive chemotherapy are the only widely available options of treatment for osteosarcoma (1). No definite therapy is yet available for this devastating disease as the molecular mechanism associated with osteosarcoma is not fully understood.
Autophagy is a crucial catabolic process in cells for maintaining homeostasis and survival through the removal and recycling of unwanted cellular materials (3). Autophagy is an evolutionarily conserved event in cells that is induced by various types of stress. It is a multi-step process involving several signal pathways. Autophagy has been shown to be pro- or anti-tumorigenic in cancer cells. Published reports indicate that autophagy may contribute to the survival of established tumors by facilitating chemoresistance during cancer therapy (4,5). This resistance to treatment is associated with the induction of autophagy in osteosarcoma and other cancers. On the contrary, autophagy mediates the cytotoxic effects of numerous anticancer and chemotherapeutic agents suggesting that autophagy may serve as a tumor suppressor event (6).
Several protein markers that are associated with autophagy have been identified. The major marker of autophagy is microtubule-associated protein light chain LC3 (7). Lipidated LC3 incorporates into the autophagosomal membrane and facilitates detection. Another marker that has been implicated widely with the induction of autophagy is P62 which is also known as Sequestosome1 (8). P62 delivers the cargo to autophagsomes by binding to the cargo on one end and to LC3 on the other end. Another protein that is upregulated during autophagy is Beclin1, which has proven to play a critical role in tumorigenesis and autophagy (9). Although the induction of autophagy during the progression of cancer has been demonstrated in numerous cancers (10,11), it is worthy to note that there is little knowledge available with regard to studies on determination of autophagy markers in osteosarcoma. In this report, to understand the significance of autophagy during the development of osteosarcoma, we have determined the expression of autophagy markers and the mechanism of autophagy in osteosarcoma patient tissues.
Methods
Tissue microarrays (TMAs)
Selected formalin fixed paraffin embedded (FFPE) tissue blocks were retrieved through the Institute Review Board (IRB)-approved protocol from the archives of the Mayo Clinic and used for preparing TMAs. Prior to use of the tissues, written informed consent was obtained from the patient for research and publication of this study. The histological diagnosis of the tissues was confirmed by a board-certified musculoskeletal pathologist at Mayo Clinic. The TMA which included 24 normal and 24 osteosarcoma tissue cores was constructed using a Galileo CK4500 semi-automated instrument in the Pathology Research Core at Mayo Clinic. Immunochemical staining was carried out with 5 micron sections using the following antibodies: anti-LC3B (1:500 dilution) (Cell Signaling Technology, Danvers, MA) anti-p62 (1:400 dilution) (Medical & Biological Laboratories, Woburn, MA), anti-PKR (1:200 dilution) and anti-phosphoPKR (1:100 dilution) (Abcam, Cambridge, MA) antibodies. A second set of TMAs representing 35 tumors and 35 control tissues were purchased from US Biomax, Inc. (Rockville, MD) and analyzed by immunostaining as described above using anti-LC3B antibodies. The quantitation of signals was carried out using BIOQUANT Image analysis system (Bioquant Image Analysis Corporation, Nashville, TN) as described (12). The average densities for protein staining were determined in normal and osteosarcoma tissues by calculating the intensities of all significant pixels in the object and dividing that value by the number of pixels as described in the manufacturer’s protocol (Bio quant Image Analysis Corporation).
Western blot hybridization
Osteosarcoma tissues and adjacent normal tissues (10 pairs) were obtained by surgical resection through the Mayo Clinic IRB-approved protocol. Normal and tumor tissues obtained were homogenized in lysis buffer and the protein concentration was determined as described (13). Cytoplasmic extract containing protein (100 µg) was analyzed by western blot hybridization using, anti-LC3 (Cell Signaling Technology) and anti-glyceraldehyde 3-phosphate dehydrogenase(anti-GAPDH) antibodies (Sigma, St. Louis, MO) as previously described (14,15). The protein levels were determined by quantitative analysis using densitometer and Imagelab software (Biorad, Hercules, CA).
Statistical analysis
The values were expressed as means ± standard error. Samples were analyzed using a Wilcoxon signed rank test for matched pairs to test the difference of the means, and the likelihood ratio was used to test difference in probabilities of differential regulation. P≤0.05 was considered statistically significant.
Results
Detection of autophagy markers by immunohistochemical staining
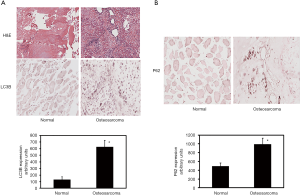
To determine whether autophagy is induced in osteosarcoma patients, we have evaluated the expression of autophagy marker LC3 in osteosarcoma and control tissues. During the induction of autophagy, LC3 undergoes proteolytic cleavage and gets converted to LC3II from its precursor form LC3I. Thus, the LC3II that is associated with autophagic flux has been detected previously by LC3 immunohistochemistry. Immunohistochemical analyses of the tissue sections reveal that the autophagic marker LC3 level is increased by 4.8-fold in osteosarcoma compared to normal tissues (Figure 1A). The increase is statistically significant. Additional investigation of TMA shows that P62 protein levels are increased in osteosarcoma patients (Figure 1B). Quantitation of immunohistochemical staining reveals a significant 2-fold increase of P62 levels.
Detection of autophagy markers by western blot analysis
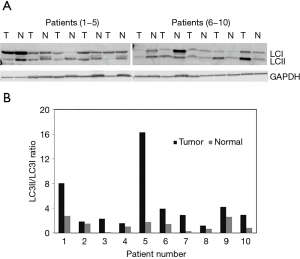
It has been previously shown using the western blot analysis that during autophagy induction the level of LC3II, the membrane bound version of LC3 protein is increased. We have performed western blot analysis using osteosarcoma tumor and adjacent normal tissues from 10 osteosarcoma patients. Normalization of the westerns with GAPDH expression levels results show that the LC3II to LC3I ratios were increased in osteosarcoma compared to the normal (Figure 2). Quantitation of western signals reveals that the LC3II levels increased in ranges from 2- to 35-fold in osteosarcoma tissue specimens.
Verification using additional set of normal and tumor tissues
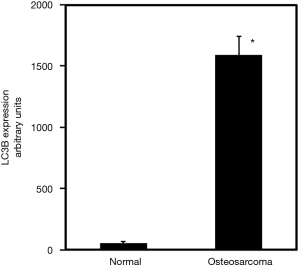
To further verify the immunohistochemistry data, from in-house samples, we examined commercial TMAs to measure LC3 expression in osteosarcoma patients. Our findings, using commercial array, corroborate the studies carried out using tissues collected at the Mayo Clinic and show that LC3 expression is increased by 29-fold in osteosarcoma compared to normal (Figure 3).
PKR activity and expression in osteosarcoma tissues
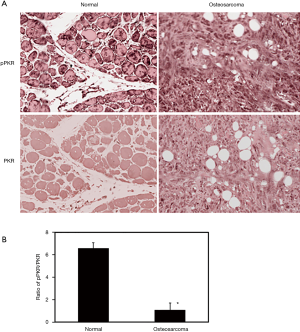
It has been shown that PKR protein is involved in the induction of autophagy in osteosarcoma cells. In order to determine if PKR plays a role in the pathological regulation of autophagy in osteosarcoma, we have determined the levels of total PKR and phosphorylated PKR (activated form) in osteosarcoma patients using TMA (Figure 4A). To statistically compare the PKR activities in osteosarcoma and normal samples, we have determined the ratio of phosphorylated PKR to total PKR. Our results illustrate that the ratio of phosphorylated PKR to total PKR is decreased by 6.6-fold in osteosarcoma patients compared to normal (Figure 4B).
Discussion
Autophagy is a regulatory event that occurs in normal and cancer cells involving the removal of organelles and cytosolic components. In this report, we have investigated the markers and mechanism of autophagy in osteosarcoma patients. Using tissue-based analysis and multiple methods, we have demonstrated that the levels of autophagy markers are increased. The results reveal that LC3 and P62 proteins are upregulated and the increase in expression of these markers correlates with the decreased activity of PKR in osteosarcoma tissues. Thus, this demonstrates a valid method for detecting the markers and regulators of autophagy which could be expanded to additional applications in clinical studies and treatments.
The dysregulation of autophagy has been implicated in several cancers (6,8,10,14). The markers of autophagy have been shown to be regulated during autophagic flux and the induction of autophagy (7,14). The autophagosome-associated protein, LC3 is incorporated into the inner and outer membrane layers of autophagosomes and thereby allows detection of autophagy at various states. P62 is an adaptor protein that plays a key role likening proteins to the surface of autophagosomes and is frequently upregulated in many tumors (6). Autophagy markers have been detected during tumor progression in many cancers but only a few have shown the expression and correlation of autophagy markers levels in osteosarcoma. Current findings demonstrate the feasibility of assessing the markers of autophagy in osteosarcoma and paired control specimen. Our results highlight that the LC3 increase is accompanied by increases in P62 and the Becklin1 proteins are increased in osteosarcoma patients (unpublished observations). These increases observed through immunohistochemical staining with LC3 and P62 are statistically significant. Furthermore, the LC3 increase observed in osteosarcoma specimen from the Mayo Clinic tissue registry is corroborated by studies involving commercial TMAs of osteosarcoma and paired controls.
It has been demonstrated that one of the hallmarks of autophagy is the conversion of the marker protein LC3I to LC3II (7). Our findings, using western blot analysis of osteosarcoma and adjacent normal tissues reveal that LC3I to LC3II protein conversion is taking place at a higher level in osteosarcoma tissues compared to normal. Thus, these verifications using control and tumor tissues from the same patients suggest that increased autophagy may contribute to tumor cell survival in osteosarcoma patients. Besides contributing to cell survival, induction of autophagy has also been shown to be associated with tumor cell migration, invasion and metastasis (16). Additional investigation is needed to understand the role of autophagy in osteosarcoma metastasis.
Targeting autophagy as treatment alone and in combination with other chemotherapeutic agents has been recently attempted in clinical trials (10). In many malignant tumors including osteosarcoma, autophagy has been shown to be induced following chemotherapy and radiotherapy. Anticancer agents that include 2-methoxyestradiol, tyrosine kinase inhibitor imatinib, the anti-epidermal growth factor receptor, proteasome inhibitors, and HDAC inhibitors have been shown to induce autophagy in a number of cancer models and clinical trials (14,17,18). While the cytoprotective role of autophagy has been addressed in these treatments, their roles in cell death are not clearly outlined. Hence, the implication of promoting autophagy in tumor cells by these agents has not been fully understood. Although several reports show the induction of autophagy during anti-tumor actions of cytotoxic drugs and chemotherapy in patients, it is yet to be uncovered whether autophagy acts as a tumor suppressor or tumor inducer. Various investigations suggest that autophagy has a context-dependent role which is dependent on cell type and stages of malignancy (19).
PKR protein is involved in the anti-tumor and anti-proliferative effects. We have shown that PKR is involved in the anti-tumor effects and induces cell death in osteosarcoma in vitro (20). Our work revealed that PKR protein is essential for anti-growth and anti-cellular activities, in addition to the induction of autophagy (14,20). Here, we show that the active PKR levels are decreased in osteosarcoma patients as shown by the decreases in the ratio of phosphorylated PKR to total PKR. The results indicate that the decreases in PKR activities correlate with the increases in autophagy markers in osteosarcoma patients. Thus, our current results corroborate the reports from our in vitro studies that PKR activity is essential for tumor suppression in osteosarcoma cells. It has been reported that tumor suppressors act as autophagy inducers and oncogenes function as inhibitors (6). Current observations contradict this phenomenon and show that the activity of the tumor suppressor PKR is decreased with the induction of autophagy in osteosarcoma tissues. This implies that PKR-role in autophagy induction is possibly associated with the effects of cytotoxic agents in osteosarcoma. Additional studies will be required to further confirm the role of PKR in autophagy during the progression of osteosarcoma disease.
In summary, we have evaluated the markers of autophagy using multiple methods in osteosarcoma patients. Studies using 69 osteosarcoma and control specimen show that markers of autophagy are increased in osteosarcoma patients indicating the induction autophagy during the progression of osteosarcoma. In addition, we show that PKR protein is associated with the pathological regulation of osteosarcoma. Future work on autophagy markers using various types and stages of osteosarcoma tissues along with the assessments on the molecular pathways that delineate cytotoxic agent-induced autophagy and cell death may improve our understanding of autophagy, which will eventually assist in the development of a new combination therapy for osteosarcoma.
Acknowledgments
The authors thank the Mayo Clinic Biomaterials and Histomorphometry Core Laboratory for tissue processing and sectioning and Ms. Suzanne Glass and Mr. James Herrick for critical reading of the manuscript.
Funding: Dr. Yuqing Li was supported by Qingdao Municipal Health Leading Academic Discipline Program QDZDXK-A-2017005. This work was supported by funding from the Mayo Clinic and the Rein in Sarcoma Foundation.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aoj.2020.02.07). AM serves as an unpaid editorial board member of Annals of Joint from September 2018 to August 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by institutional review board (IRB) of Mayo Clinic (No. 11-004649 and 15-005628) and written informed consent was obtained from the patients for publication of the results and accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arndt CA, Rose PS, Folpe AL, et al. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin Proc 2012;87:475-87. [Crossref] [PubMed]
- Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol 2010;8:705-18. [PubMed]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev cell 2004;6:463-77. [Crossref] [PubMed]
- Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913-30. [Crossref] [PubMed]
- Reumann S, Shogren KL, Yaszemski MJ, et al. Inhibition of Autophagy Increases 2-Methoxyestradiol-Induced Cytotoxicity in SW1353 Chondrosarcoma Cells. J Cell Biochem 2016;117:751-9. [Crossref] [PubMed]
- Morselli E, Galluzzi L, Kepp O, et al. Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta 2009;1793:1524-32. [Crossref] [PubMed]
- Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008;4:151-75. [Crossref] [PubMed]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009;137:1001-4. [Crossref] [PubMed]
- Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009;11:468-76. [Crossref] [PubMed]
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019;9:1167-81. [Crossref] [PubMed]
- Yang ZJ, Chee CE, Huang S, et al. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011;10:1533-41. [Crossref] [PubMed]
- Bravo D, Shogren KL, Zuo D, et al. 2-Methoxyestradiol-Mediated Induction of Frzb Contributes to Cell Death and Autophagy in MG63 Osteosarcoma Cells. J Cell Biochem 2017;118:1497-504. [Crossref] [PubMed]
- Bravo D, Salduz A, Shogren KL, et al. Decreased local and systemic levels of sFRP3 protein in osteosarcoma patients. Gene 2018;674:1-7. [Crossref] [PubMed]
- Yang C, Shogren KL, Goyal R, et al. RNA-dependent protein kinase is essential for 2-methoxyestradiol-induced autophagy in osteosarcoma cells. PLoS One 2013;8:e59406 [Crossref] [PubMed]
- Zuo D, Shogren KL, Zang J, et al. Inhibition of STAT3 blocks protein synthesis and tumor metastasis in osteosarcoma cells. J Exp Clin Cancer Res 2018;37:244. [Crossref] [PubMed]
- Sharifi MN, Mowers EE, Drake LE, et al. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep 2016;15:1660-72. [Crossref] [PubMed]
- Chude CI, Amaravadi RK. Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors. Int J Mol Sci 2017; [Crossref] [PubMed]
- Kwon Y, Kim M, Jung HS, et al. Targeting Autophagy for Overcoming Resistance to Anti-EGFR Treatments. Cancers (Basel) 2019; [Crossref] [PubMed]
- Kondo Y, Kanzawa T, Sawaya R, et al. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005;5:726-34. [Crossref] [PubMed]
- Shogren KL, Turner RT, Yaszemski MJ, et al. Double-stranded RNA-dependent protein kinase is involved in 2-methoxyestradiol-mediated cell death of osteosarcoma cells. J Bone Miner Res 2007;22:29-36. [Crossref] [PubMed]
Cite this article as: Ji Y, Okuno MN, Shogren KL, Fritchie K, Okuno SH, Yaszemski MJ, Maran A. Autophagy markers and RNA-dependent protein kinase (PKR) activity in osteosarcoma diagnosis and treatment. Ann Joint 2020;5:26.


